In the era of interdisciplinary research, molecular dynamics (MD) simulation has become an indispensable tool in fields such as chemistry, biomedicine, and materials science. Against this backdrop, Professor Yi Qin Gao's team from the School of Chemistry and Molecular Engineering at Peking University, Yi Isaac Yang's group from the Institute of Systems and Physical Biology at Shenzhen Bay Laboratory, and Paratera have successfully completed the cloud deployment of the domestically developed molecular dynamics simulation engine SPONGE, marking a significant breakthrough for China in the field of autonomous and controllable high-performance computing and molecular simulation.
In this collaboration, Peking University and Shenzhen Bay Laboratory lead the core algorithm development of SPONGE, leveraging innovative GPU acceleration technology and AI-driven enhanced sampling methods (such as metadynamics and deep-learning-based enhanced sampling), significantly improving simulation accuracy and speed; Paratera, relying on its cloud computing power platform, providing high-performance computing resources to support SPONGE, enabling it to efficiently process ultra-large-scale molecular systems (such as protein-ligand interactions and molecular dynamics at the surface of nanomaterials) while supporting multi-task parallel computing, significantly reducing research costs and time.
This collaboration, through open-source technology and modular design, provides flexible and scalable solutions for interdisciplinary research.

Step 1: Paratera Cloud Desktop Login Interface

Step 2: Click SPONGE
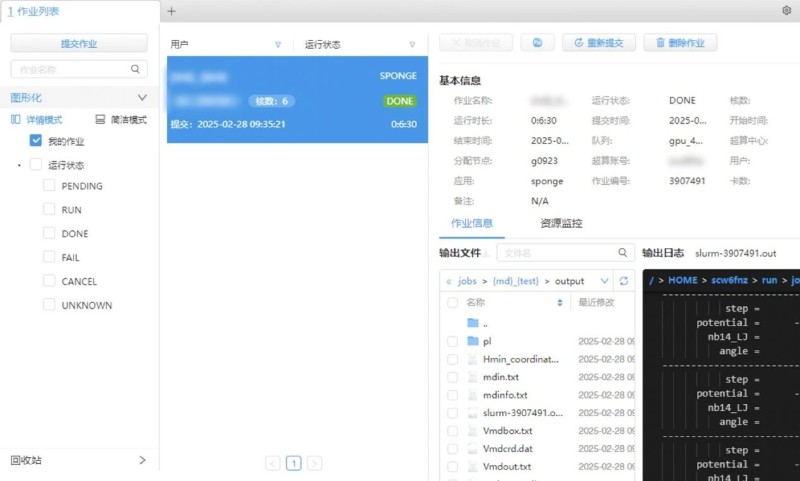
Step 3: Perform relevant operations in the SPONGE interface
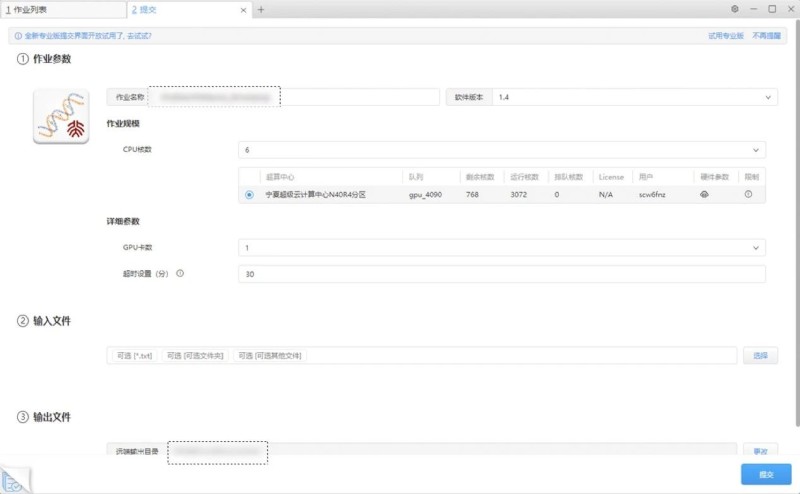
Step 4: Select the scope of work and submit
User benefits: Empowering the entire chain from research to industry
1. Efficiency leap:
SPONGE's GPU acceleration technology increases simulation speed by more than 10 times. Combined with cloud-based elastic computing power, users can quickly complete computing tasks that traditionally take weeks, especially in time-sensitive fields such as drug discovery and new material design.
2. Precision and functional expansion:
SPONGE matches the accuracy of leading international software (such as Amber) and supports customized simulations of complex systems (such as biomolecules and special material surfaces), meeting the needs of cutting-edge research.
3. Cost Optimization:
Cloud deployment eliminates the need for local high-performance hardware investments, and the pay-as-you-go model lowers the barrier to entry for small and medium-sized teams, promoting inclusive research.
4. Open Ecosystem:
Modular design (CudaSPONGE, MindSPONGE, Xponge) supports seamless integration with deep learning and molecular docking tools (such as DSDP), driving the deep integration of AI for Science.
Case Studies: SPONGE's Diverse Application Scenarios
- Biomedical: Precise simulation of protein conformational changes to accelerate targeted drug screening;
- Materials Science: Analyzing the dynamic behavior of nanomaterial surfaces to guide the development of high-performance materials;
- Education: Open-source features support university teaching and research training, fostering interdisciplinary talent.
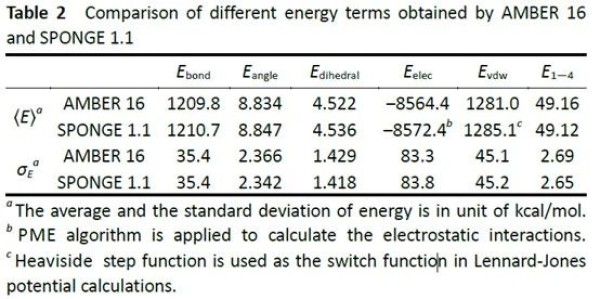
SPONGE shows no significant difference in computational accuracy compared to Amber
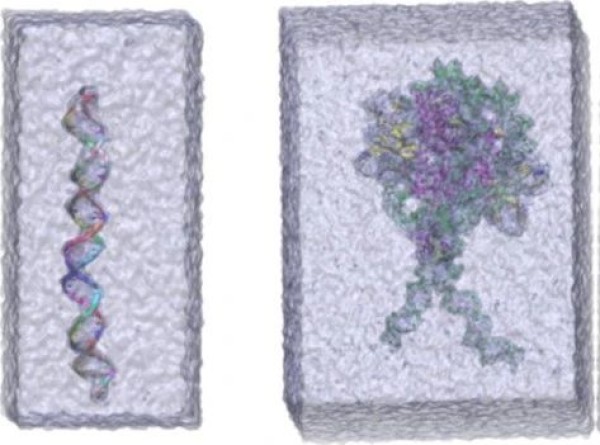
SPONGE performs well in simulating biological macromolecular systems
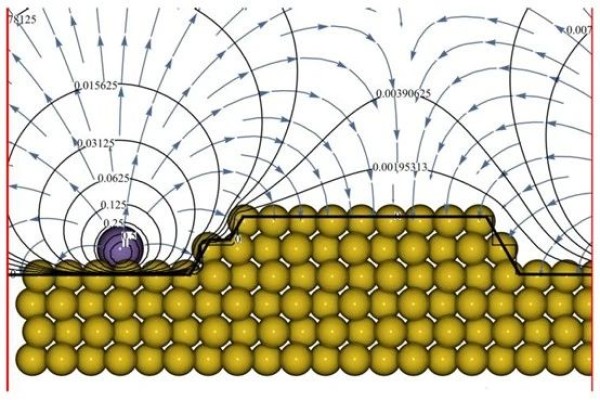
SPONGE can even be used for customized calculations of surface information for some special material systems
References:
1. Huang Y. P. et al., *Chin. J. Chem.* 2022, DOI:10.1002/cjoc.202100456
2. Xia Y. et al., *J. Open Source Softw.* 2022, DOI:10.21105/joss.04467
